Prostate cancer (PCa) is the second most common cancer in men with an estimated prevalence of 1.1 million worldwide in 2012. This heterogeneous disease resulted in approximately 307,000 deaths, making it the fifth leading cause of cancer-related death in men worldwide [1]. The incidence of PCa varies more than 25-fold worldwide in part due to different practices of prostate-specific antigen (PSA) testing and subsequent prostate biopsy (PBx).
The diagnosis and staging of PCa is based on digital rectal examination (DRE), PSA blood levels, imaging (multi-parametric (mp) MRI, cross-sectional abdominopelvic CT and bone scan) and histological verification by PBx. Historically, a transrectal ultrasound-guided PBx (TRUS-PBx) was offered as a result of a raised age-specific PSA or suspicious findings on DRE [2]. However, TRUS-PBx is an invasive procedure associated with risks including infection, haematuria, haematospermia and pain [3].
Over 100,000 TRUS-PBx are performed annually in the UK and the incidence of detecting PCa in men with a PSA between 4 and 10ng/ml is around 30%, and around 15% of PCa detected by TRUS-PBx have a PSA level below 4ng/ml [4]. Men who have an initial negative PBx but have a rising PSA are subject to repeat PBx (rPBx). This automatic approach may increase healthcare costs and put patients at risk of PBx complications. In addition, there is a risk of over-diagnosing and over-treating clinically insignificant PCa.
A randomised-controlled trial (RCT) of 731 men did not show any mortality differences at 10 years between radical prostatectomy (RP) and watchful waiting (WW) in screen-detected clinically organ-confined PCa [5]. The world’s largest RCT on RP versus radical radiotherapy versus active monitoring also did not show any survival benefits within the three groups at 10 years [6]. Therefore, treating low-risk organ-confined localised disease may not be necessary.
What is important is predicting PBx outcomes and identifying men at risk of significant high-grade disease (Gleason score 4+3=7 and above) in order to reduce the number of unnecessary PBx and to allow a more personalised PBx and treatment decision. Prostate biopsy decision-making is difficult and there are different biomarkers, imaging adjuncts and nonograms / risk calculators that may help improve diagnosis of significant disease on initial PBx and predict outcomes of rPBx in men with negative initial PBx.
Current European guidelines suggest that one set of rPBx is warranted where there is a suspicious DRE, rising / persistently elevated PSA value and a histopathological finding suggestive of potential malignancy (atypical small acinar proliferation – ASAP, high-grade prostatic intraepithelial neoplasia – HGPIN, intraductal carcinoma) on initial biopsy. There are no further recommendations on subsequent rPBx [2].
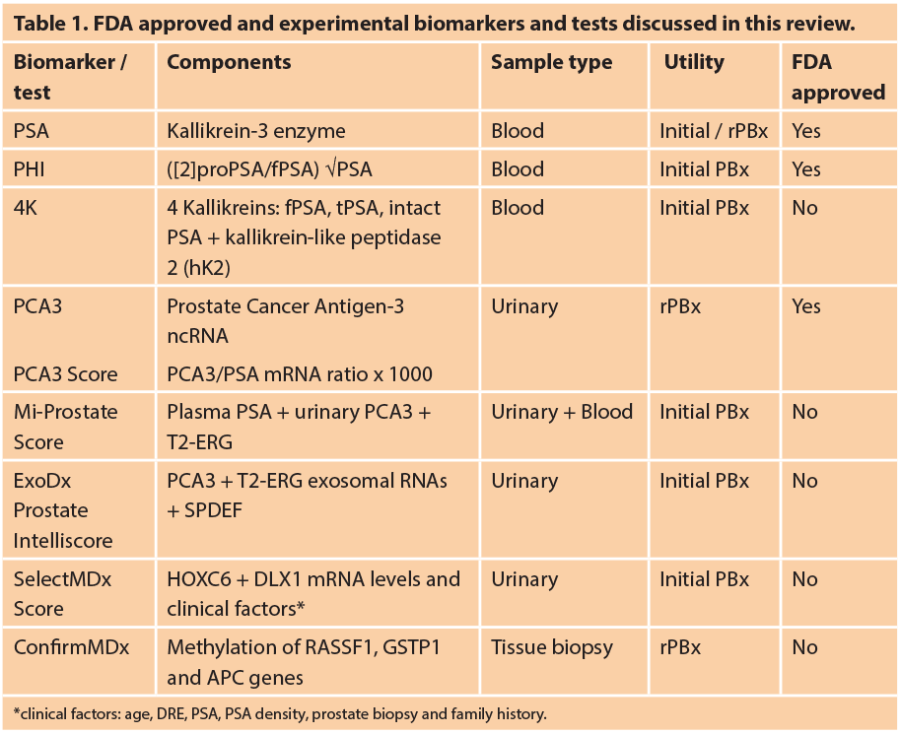
Over the past two decades, there have been promising discoveries of biomarkers and molecular and clinical scores to improve PCa detection, stratify patient for rPBx and to predict biochemical recurrence following radical treatment. The aim of the current review is to discuss current US Food and Drug Administration (FDA) approved and experimental biomarkers and tests for PBx decision (Table 1).
Prostate-specific antigen
Prostate-specific antigen, also known as kallikrein-3 (KLK3), is a glycoprotein enzyme encoded in humans by the KLK3 gene. PSA is present in the blood in multiple forms known as isoforms, and some of these forms are more cancer-specific. PSA testing was introduced in the late 1980s to aid PCa diagnosis. Whilst PSA is relatively organ-specific, elevations in serum PSA levels are not cancer-specific.
The threshold for an abnormal PSA level has been 4.0ng/ml and the associated estimated sensitivity and specificity are 21-44% and 91-92% respectively. Moreover, many men may have PCa despite having a low PSA level. Free/total (f/t) PSA ratio may be used to differentiate benign prostate hyperplasia (BPH) from PCa. It stratifies the PCa risk with 4-10ng/ml total PSA and negative DRE. In a reported study, 56% of men with f/t PSA <0.10 were found to have PCa on PBx, in contrast only 8% of men with f/t PSA >0.25ng/ml were found to have PCa [7].
A few tests measuring a range of KLK in serum and plasma are now commercially available, including the FDA-approved Prostate Health Index (PHI) test and the four kallikrein score (4K). Both the PHI and the 4K score are used with the intention of reducing the number of unnecessary PBx [8,9].
Prostate Health Index
The PHI blood test combines fPSA and tPSA and the [-2]pro-PSA isoform (p2PSA) and is calculated using the following formula ([−2]pro-PSA/fPSA) × √PSA. Isoforms of PSA have similar biological roles but differ in sequence and structural form. PHI was approved by the FDA in 2012 for men with PSA levels between 4 and 10ng/ml. PHI can improve the detection of high-risk PCa and is associated with PCa aggressiveness [10].
Many prospective trials have shown that PHI outperforms its individual components of total, free and [-2]pro-PSA for detecting clinically significant PCa and reducing the number of unnecessary PBx (11–15) [11-15]. A recent systematic review (SR) showed an additional value by the increase in area under the curve (AUC) value by 2-11% [16]. The PHI has also been shown to add predictive value to mpMRI in detecting significant PCa in a rPBx population [17].
4K score
The 4K score measures four kallikrein markers, fPSA, tPSA, intact PSA and kallikrein-like peptidase 2 (hK2). A prospective US study enrolled 1012 men and showed that the 4K score demonstrated excellent diagnostic performance in detecting significant PCa (AUC 0.82) with a possible reduction of 30-58% in the number of PBx [18]. A meta-analysis analysed 8500 men and showed that the 4K score is associated with an improvement of 8-10% in predicting biopsy-confirmed PCa with an estimate of avoiding 48-56% of current PBx [19]. However, unlike PHI, the 4K score is not yet FDA approved. When comparing 4K with PHI, a study of 531 men found similar performances in both tests in detecting significant PCa (AUC 4K, 71.8 vs. PHI, 71.1) and reducing the number of unnecessary PBx [20].
The PHI and 4K score measure only a few known isoforms of PSA that give meaningful information if they are present at a given time. IsoPSA is a structure-based (rather than concentration-based) test that incorporates the entire spectrum of PSA structural changes. IsoPSA has been shown in a prospective study to be more superior than standard PSA at predicting PCa (versus benign) and high-grade PCa (versus low-grade and benign). Once validated, IsoPSA may be another potential assay that would help to better select at risk men for PBx [21].
Prostate cancer antigen 3
PCA3 is a long non-coding RNA discovered in 1999 and was FDA approved in 2012 [22,23]. It has been shown to be useful when used in conjunction with PSA (PCA3 score, PCA3/PSA mRNA ratio x 1000) to predict PCa on rPBx. Unlike PSA, PCA3 is organ and PCa-specific. It can be detected in urinary samples (PROGENSA® assays), more readily after DRE and has a sensitivity of ~67% and a specificity of ~83%. A European prospective multicentre study [24] and the REDUCE RCT [25] demonstrated that the PCA3 score is higher in men with a positive biopsy than in men with a negative biopsy. PCA3 score increases with PCa volume, however, there are conflicting data about whether it independently predicts the Gleason score. When comparing the diagnostic accuracy of PCA3 with PHI to predict PCa in men undergoing initial PBx, PCA3 performed better at diagnosing overall PCa (AUC 0.71 vs. PHI, 0.65), but less so at detecting significant PCa (AUC 0.55 vs. PHI, 0.80) [26]. Currently, the main use of the PROGENSA® test is to aid rPBx decisions, but its clinical effectiveness for this purpose is uncertain [27].
Mi-Prostate Score Gene
rearrangements have been described in multiple cancers. The TMPRSS2-ERG fusion gene comprises the androgen-responsive genes TMPRSS2 (transmembrane protease, serine 2) and erythroblast transformation-specific (ETS)-related genes (ERG), and was observed in ~40-80% of PCa in 2005 [28]. Both genes are located on chromosome 21, and the TMPRSS2-ERG score is calculated using the formula (TMPRSS2-ERG mRNA/PSA RNA copies) x 100,000. The Mi-Prostate Score (MiPS) combines the prognostic significance of urinary TMPRSS2-ERG (T2:ERG) and urinary PCA3 with serum PSA to generate a PCa risk assessment score. Although not yet FDA approved, the MiPS has been shown to be more superior to PSA alone in predicting biopsy-confirmed PCa and high-grade disease in a validation cohort of 1225 samples. The AUC for MiPS at predicting high-grade PCa was 0.75; vs. PSA alone, 0.59; vs. PSA+T2:ERG, 0.69; vs. PSA+PCA3, 0.73 [29].
ExoDx
The ExoDx Prostate Intelliscore is an urinary test which measures PCA3 and T2-ERG exosomal RNA levels, normalised with SAM pointed domain-containing ETS transcription factor [30]. The test was validated on 195 urinary samples and showed good clinical performance in predicting high-grade PCa with a NPV of 97.5% [31]. Recently, a larger validation cohort of 519 men was analysed and combining the ExoDx test with clinical ‘standard of care’ (SOC) risk factors such as PSA, age, race and family history resulted in improved prediction of high-grade PCa compared to SOC alone (AUC, 0.73 vs. SOC, 0.63). Using a predefined cut point, 27% PBx would have been avoided, missing 5% of patients with high-grade Gleason 4+3=7 PCa [32].
“Prostate biopsy decision-making is difficult and there are different biomarkers, imaging adjuncts and nonograms / risk calculators that may help improve diagnosis of significant disease on initial PBx and predict outcomes of rPBx in men with negative initial PBx.”
SelectMDx
The SelectMDx test is based on urinary mRNA biomarker. HOXC6 and DLX1 mRNA levels have been shown to be good predictors for the detection of high-grade PCa. Risk scores combining HOXC6 and DLX1 mRNA levels with clinical risk factors such as age, DRE, PSA, PSA density, prostate biopsy and family history are able to accurately detect high-grade clinically significant PCa. Using a validation cohort of 386 urinary samples, SelectMDx had an AUC of 0.90 for predicting high-grade PCa. Choosing a cut-off with a negative predictive value (NPV) of 98% for high-grade PCa, a reduction of PBx by 42% and a reduction of the unnecessary PBx by 53% were obtained [33].
ConfirmMDx
Epigenetic events such as DNA methylation, histone modifications and microRNA (miRNA) regulation have been well studied in PCa [34]. The ConfirmMDx test is a tissue-based epigenetic test that quantifies the methylation level of promoter regions of three genes (RASSF1, GSTP1 and APC) in benign prostatic tissue [35]. The MATLOC study analysed archived negative initial PBx samples from 498 men and showed a NPV of 90% [36]. The DOCUMENT multicentre USA trial also analysed archived negative PBx samples from 350 men and showed a similar NPV of 88% [37].
A multicentre study showed a negative predictive value of 88% when methylation was absent in all three markers, suggesting that rPBx could be avoided in these men [36,38]. Given the limited currently available data, no recommendation can be made regarding its routine application.
Short and microRNAs
Evolving information on the roles of microRNAs (miRNAs) has established them as potential biomarkers and therapeutic targets. They are utilised as urinary markers for diagnosing urological cancers and appear to show great potential in managing cancers [39,40]. PCA3 is not used within the UK National Health System due to its unclear biological function and high cost in preventing degradation prior to laboratory evaluation. Short or microRNAs appear to be promising biomarkers as their small size protects them from endogenous RNase degradation. A short-RNA within PCA3 termed PCA3-shRNA has been discovered and is expressed in both urinary and PBx specimens. This short RNA is overexpressed in PCa urinary samples when compared with BPH samples and appears to function by regulating mRNA through complementary base binding. With more validation, PCA3-shRNA may replace the current longer PCA3 assay [41,42].
Many studies report numerous upregulated and downregulated miRNAs, with conflicting data. This reflects different profiling strategies, differences in analytical thresholds, study design (samples and methods) and disease heterogeneity. miRNA expression is variable and differs according to the phases of development (initiation, progression or metastasis) or treatment exposure (ADT, radiotherapy, chemotherapy). Many studies have looked into miRNA as diagnostic and prognostic biomarkers. Fabris et al. conducted a systematic review and highlighted plasma miRNAs that were consistently altered in PCa with diagnostic properties. These miRNAs include upregulatory, miR-141, 375, 221, 21; and downregulatory, miR-181a [43].
Conclusion
Prostate cancer is a heterogeneous disease and over the past decade many biomarkers, tests and risk scores have been developed to aid initial and rPBx decisions in order to reduce over-diagnosis and over-treatment of clinically insignificant disease, and to identify high-grade clinically significant disease to allow personalised treatment decisions. A test or scoring system through a multi-modal approach incorporating molecular markers, imaging (mpMRI) and clinical risk parameters appears to be more appropriate in optimising personalised PBx decisions.
TAKE HOME MESSAGE
-
Prostate cancer is a heterogeneous disease; currently a diagnostic approach based on raised PSA leads to over-diagnosis and over-treatment of insignificant disease.
-
A more precise diagnostic approach incorporating molecular biomarkers, imaging and clinical risk parameters is needed to identify clinically significant PCa, reduce the number of unnecessary PBx and allow a more personalised treatment decision.
-
Currently FDA approved tests include PROGENSA® PCA3 score and PHI.
-
Other clinical and experimental biomarkers / tests that have been evaluated but not FDA approved include 4K, Mi-prostate score, ExoDx Prostate Intelliscore, SelectMDx score, ConfirmMDx and miRNAs.
References
1. International Agency for Research on Cancer. GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. 2012;
http://globocan.iarc.fr/
Pages/fact_sheets_cancer.aspx
2. Mottet N, Bellmunt J, Briers E, et al. EAU-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer. Eur Urol 2017;71(4):618-29.
3. Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol 2013;64(6):876-92.
4. Thompson IM, Pauler DK, Goodman PJ, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level ≤4.0 ng per milliliter. N Engl J Med 2004;350(22):2239-46.
5. Wilt TJ. The Prostate Cancer Intervention Versus Observation Trial: VA/NCI/AHRQ Cooperative Studies Program #407 (PIVOT): design and baseline results of a randomized controlled trial comparing radical prostatectomy with watchful waiting for men with clinically localized prostate cancer. J Natl Cancer Inst Monogr 2012;2012(45):184-90.
6. Hamdy FC, Donovan JL, Lane JA, et al. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med 2016;375(15):1415-24.
7. Catalona W, Partin A, Slawin K, Al E. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: A prospective multicenter clinical trial. JAMA 1998;279(19):1542-7.
8. Loeb S, Catalona WJ. The Prostate Health Index: a new test for the detection of prostate cancer. Ther Adv Urol 2014;6(2):74-7.
9. Bryant RJ, Sjoberg DD, Vickers AJ, et al. Predicting high-grade cancer at ten-core prostate biopsy using four kallikrein markers measured in blood in the ProtecT Study. J Natl Cancer Inst 2015;107(7):djv095.
10. Lazzeri M, Haese A, Abrate A, et al. Clinical performance of serum prostate-specific antigen isoform [-2]proPSA (p2PSA) and its derivatives, %p2PSA and the prostate health index (PHI), in men with a family history of prostate cancer: results from a multicentre European study, the PROMEtheuS. BJU Int 2013;112(3):313-21.
11. Tosoian JJ, Druskin SC, Andreas D, et al. Use of the Prostate Health Index for detection of prostate cancer: results from a large academic practice. Prostate Cancer Prostatic Dis 2017;20(2):228-33.
12. White J, Shenoy BV, Tutrone RF, et al. Clinical utility of the Prostate Health Index (phi) for biopsy decision management in a large group urology practice setting. Prostate Cancer Prostatic Dis 2018;21(1):78-84.
13. Loeb S, Sanda MG, Broyles DL, et al. The Prostate Health Index selectively identifies clinically significant prostate cancer. J Urol 2015;193(4):1163-9.
14. Wang W, Wang M, Wang L, et al. Diagnostic ability of %p2PSA and prostate health index for aggressive prostate cancer: a meta-analysis. Sci Rep 2014;4(1):5012.
15. Catalona WJ, Partin AW, Sanda MG, et al. A multicenter study of [-2]Pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J Urol 2011;185(5):1650-5.
16. Lamy P-J, Allory Y, Gauchez A-S, et al. Prognostic biomarkers used for localised prostate cancer management: a systematic review. Eur Urol Focus 2017; [Epub ahead of print].
17. Gnanapragasam VJ, Burling K, George A, et al. The Prostate Health Index adds predictive value to multi-parametric MRI in detecting significant prostate cancers in a repeat biopsy population. Sci Rep 2016;6(1):35364.
18. Parekh DJ, Punnen S, Sjoberg DD, et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur Urol 2015;68(3):464-70.
19. Voigt JD, Zappala SM, Vaughan ED, Wein AJ. The Kallikrein Panel for prostate cancer screening: Its economic impact. Prostate 2014;74(3):250-9.
20. Nordström T, Vickers A, Assel M, et al. Comparison between the Four-kallikrein Panel and Prostate Health Index for predicting prostate cancer. Eur Urol 2015;68(1):139-46.
21. Klein EA, Chait A, Hafron JM, et al. The single-parameter, structure-based IsoPSA assay demonstrates improved diagnostic accuracy for detection of any prostate cancer and high-grade prostate cancer compared to a concentration-based assay of total prostate-specific antigen: a preliminary report. Eur Urol 2017;193(0):1163‑9.
22. Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999;59(23):5975-9.
23. Clarke RA, Zhao Z, Guo A-Y, et al. New genomic structure for prostate cancer specific gene PCA3 within BMCC1: implications for prostate cancer detection and progression. PLoS One 2009;4(3):e4995.
24. Haese A, de la Taille A, van Poppel H, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol 2008;54(5):1081-8.
25. Aubin SMJ, Reid J, Sarno MJ, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol 2010;184(5):1947-52.
26. Seisen T, Rouprêt M, Brault D, et al. Accuracy of the prostate health index versus the urinary prostate cancer antigen 3 score to predict overall and significant prostate cancer at initial biopsy. Prostate 2015;75(1):103‑11.
27. Nicholson A, Mahon J, Boland A, et al. The clinical effectiveness and cost-effectiveness of the PROGENSA® prostate cancer antigen 3 assay and the Prostate Health Index in the diagnosis of prostate cancer: a systematic review and economic evaluation. Health Technol Assess (Rockv) 2015;19(87):1-192.
28. Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310.
29. Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG Plus PCA3 for individualized prostate cancer risk assessment. Eur Urol 2016;70(1):45-53.
30. Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009;100(10):1603-7.
31. Donovan MJ, Noerholm M, Bentink S, et al. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis 2015;18(4):370-5.
32. McKiernan J, Donovan MJ, O’Neill V, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol 2016;2(7):882.
33. Van Neste L, Hendriks RJ, Dijkstra S, et al. Detection of high-grade prostate cancer using a urinary molecular biomarker–based risk score. Eur Urol 2016;70(5):740-8.
34. Jerónimo C, Bastian PJ, Bjartell A, et al. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol 2011;60(4):753-66.
35. Van Neste L, Bigley J, Toll A, et al. A tissue biopsy-based epigenetic multiplex PCR assay for prostate cancer detection. BMC Urol 2012;12(1):16.
36. Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC Study. J Urol 2013;189(3):1110-16.
37. Partin AW, Van Neste L, Klein EA, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014;192(4):1081-7.
38. Partin AW, Van Neste L, Klein EA, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014;192(4):1081-7.
39. Miah S, Pang K, Catto JWF. MicroRNA and urothelial cell carcinoma. BJU Int 2014;113(5):811-12.
40. Miah S, Catto JWF. MicroRNA in prostate cancer: an opportunity to individualize patient care. J Urol 2012;187(4):1155-6.
41. Drayton RM, Rehman I, Clarke R, et al. Identification and diagnostic performance of a small RNA within the PCA3 and BMCC1 gene locus that potentially targets mRNA. Cancer Epidemiol Biomarkers Prev 2015;24(1):268-75.
42. Pang KH, Rosario DJ, Morgan SL, Catto JWF. Evaluation of a short RNA within prostate cancer gene 3 in the predictive role for future cancer using non-malignant prostate biopsies. PLoS One 2017;12(4):e0175070.
43. Fabris L, Ceder Y, Chinnaiyan AM, et al. The potential of microRNAs as prostate cancer biomarkers. Eur Urol 2016;70(2):312-22.
Declaration of competing interests: None declared.