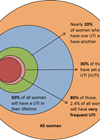
Urinary tract infection (UTI) is a common reason for seeking medical care in both primary and secondary settings. Half of women will have at least one episode of cystitis in their lifetime, and a third of them will have experienced one before 24 years old of age [1]. In England, lower urinary tract infections accounted for 22% (equivalent to seven million prescriptions) of the total antibiotics prescribed in the 2019/20 period, and 41% of those were prescribed to people over 70 [2].
Current standard diagnostic techniques
The clinical diagnosis of a UTI can be established through a targeted patient history and evaluation of the presenting symptoms. According to the most recent guidance from the National Institute for Health and Care Excellence (NICE), it is recommended to initiate empirical antibiotic treatment in women under 65 years old if they exhibit two or more key urinary symptoms (such as dysuria, new nocturia, or cloudy urine), without the requirement of a urine dipstick test. In addition to the urine dipstick, the ‘gold standard’ investigation is a urine culture [3].
Urine dipstick
The urine dipstick is used to detect nitrites and leucocyte esterase in urine, indicating bacteriuria and pyuria. Its diagnostic value is limited in uncomplicated cystitis but can be helpful when the diagnosis in unclear [1]. The meta-analysis study by Deville et al. reported sensitivities of 45-60% for nitrites, and 48-86% for leucocyte esterase to predict UTIs, indicating relatively low sensitivity levels [4]. Many factors can also affect the dipstick results, leading to both false positive and false negative results. For example, the non-nitrite producing organisms, such as staphylococcus and enterococcus, can result in a false-negative reaction [5]. The automated urinary flow cytometry is more precise in detecting bacteria, pyuria, red blood cells and epithelial cells.
Urine microscopy, culture, and sensitivity
Urine samples suspected of UTIs are sent to the microbiology laboratory. Initially, microscopy testing is performed to detect the presence of bacteria, which typically takes one to two hours. The subsequent identification of a pathogen can take up to 24 hours. Following identification, the sample undergoes antimicrobial susceptibility testing (AST), which can take 24 to 48 hours [5]. In cases where patients exhibit atypical symptoms, signs of upper UTIs or complicated cystitis, recurrent UTIs, or in pregnant patients, urine culture testing is advised. However, in otherwise healthy patients, empirical treatment is recommended to avoid delays [3].
The threshold for diagnosing a UTI based on urine culture is typically a bacterial count of more than 105 colony-forming units per millilitre (CFU/ml). Cultures with counts ranging from 10² to 10³ CFU/ml are generally not reported as positive in standard laboratory settings. The criterion of 105 CFU/ml originates from the original study by Kass et al. from the 1950s, who described significant bacteriuria in women with upper urinary tract infections as 105 CFU/ml [6]. Even though this has never been validated for lower urinary tract infections, it is still being used as a global standard for diagnosing UTIs. It is now recognised that 30-50% of patients with lower UTIs have less than 105 CFU/ml, which suggests some of the lower UTIs can be undertreated [7]. Additionally, certain bacteria, such as uropathogenic E. coli, can form intracellular bacterial communities, undetected by routine mid-stream urine (MSU) or catheter specimen urine (CSU) samples [8].
The asymptomatic bacteriuria can be mistaken for UTI and lead to inappropriate antibiotic prescribing. The incidence of asymptomatic UTI varies among patient populations, ranging from 5-15% in healthy individuals up to 30-60% in elderly patients residing in long-term care facilities [9]. Furthermore, the urine samples can become contaminated leading to false positive results.
Extended cultures
Extended cultures, where urine is incubated in a broth rather than on a standard agar plate, provide improved representation of pathogen growth. If no growth is observed within three days, the incubation period is extended for an additional three days. The extended culture approach enables the detection of slower-growing organisms such as anaerobic bacteria like lactobacillus and enterococcus. Additionally, it allows for the identification of fungal infections and provide information on pathogen sensitivities. While standard cultures can detect fast-growing bacteria like E. coli, slower-growing pathogens and those that thrive in unfavourable conditions may require up to five days to manifest visible growth. Some pathogens may even go undetected entirely using standard culture methods [10].
Issues with current UTI diagnostics
The diagnosis of urinary tract infections (UTIs) can present challenges in both primary and secondary care settings due to the non-specific nature of signs and symptoms. Conventional ‘gold standard’ diagnostic methods can lead to both overdiagnosis and underdiagnosis of UTIs. The culture testing is labour intensive and time consuming, not allowing the point-of-care management of the UTIs. There is a need for innovative diagnostic techniques that can enhance accurate identification of pathogens with a faster turnaround time, while also reducing the workload associated with negative sample results.
More advanced techniques
Polymerase chain reaction
Polymerase chain reaction (PCR) is a laboratory technique used to amplify targeted DNA sequences. Since its development in the 1980s, PCR has made large advances in molecular biology and genetics. PCR provides rapid identification of a specific pathogen with high sensitivity and specificity and allows for a high detection rate of a single pathogen compared to urine culture. Moreover, it holds the capability to detect multiple pathogens which standard urine cultures often fail to do. This is an advantage, as 30-40% of urinary tract infections are polymicrobial, and detection of these pathogens can aid in accurate antimicrobial treatment [11].
The real-time PCR can be used for rapid detection of pathogens and detect the presence of known antibiotic resistant genes [5]. Unfortunately, the current PCR techniques can only detect the presence of a pathogen, and not its concentration. This limitation increases the risk of detecting contaminants rather than true pathogens. Additionally, PCR cannot be used for antibiotic susceptibility testing and is more expensive than standard urine cultures [5].
In the UK, PCR diagnostic testing for UTI is only available through private laboratory clinics. Patients can request their tests online and receive a sample collection package that they require to post. As these panel tests are only able to detect a panel of commonly known pathogens, these can miss the dominant species up to 60% of the time. In addition to this, it can take up to seven days for the patients to receive their results [12].
Currently, it is yet to be determined whether the benefit of PCR diagnostic UTI testing will outweigh its financial cost and logistics.
Fluorescence in situ hybridisation (FISH)
The fluorescence in situ hybridisation (FISH) utilises fluorescence-labelled nucleic acid probes that hybridise to specific genomic sequences on chromosomes, allowing for microscopic detection. This technique is commonly employed in genetic diseases, gene mapping, and the identification of oncogenes [5].
FISH assay kits have been approved for the detection of pathogens in positive blood cultures. These assays can help to identify pathogens including S. aureus, E. faecalis, E. coli, P. aeruginosa, K. pneumoniae and multiple Candida species [5]. The rapid assays can be processed within 20 minutes with sensitivity and specificity of over 96% [5].
Like PCR, FISH is unable to incorporate antibiotic susceptibility. In a study by Wu et al., it was reported that FISH assays failed to identify some E. coli strains in urine samples. This limitation may be attributed to mutations within the RNA of the E. coli, which poses challenges in identifying mutated pathogens [13].
New developing techniques
Biosensors
A biosensor is a molecular sensing device designed to detect biological entities. It consists of a recognition element, which interacts specifically with the target analyte, and a signal transducer that converts this interaction into measurable signals. Biosensors have the capability to detect and quantify pathogens and provide information on antibiotic susceptibility [14].
There are various types of different biosensors available. For example, electrochemical biosensors use specific DNA probes to target bacterial RNA, and optical biosensors integrated with microfluidics can produce fluorescence imaging for detection of pathogens and antibiotic susceptibility [15].
Biosensors are well suited for point-of-care testing, as they provide rapid real-time and highly sensitive results, especially when combined with the microfluidics technology. They serve as miniature analytical tools and require only small sample volumes, making them user-friendly and convenient.
Further in-depth research is required into the use of biosensors, as multiple factors, including chemical reactions, electronics, nanotechnology, and biological materials, can cause interference. A major challenge lies in developing biosensors capable of detecting multiple pathogens and those with resistant strains. Additionally, the availability of UTI biosensors is currently limited.
Microfluidics
Microfluidics is a field of science and technology that focuses on creating tiny devices containing microchannels through which fluids can flow. The physics behind the behaviour of the tiny volumes of fluids (femoliters – fL) is quite different from the physical features of the everyday volumes of fluids. These unique features allow microfluidics chips to be an efficient and rapid tool in biological analysis. A well-known example of a non-medical microfluidics device is an inkjet printer, which utilises microfluidic principles to precisely deposit small droplets of ink onto a surface.
One of the most important applications of microfluidics for diagnostics are point-of-care devices, as small volumes of fluid can be processed and analysed much faster than traditional laboratory techniques. By integrating biosensors with microfluidic handling systems, complex molecular assays required for pathogen detection and antibiotic sensitivity testing can be performed within a short period of time [16].
NICE reviewed developing diagnostic techniques
A NICE committee has recently reviewed the evidence around different point-of-care tests for suspected UTIs in primary and secondary care [17]. The tests were categorised as rapid (providing results in less than 40 minutes) or culture-based (providing results in approximately 16-24 hours). Although the committee did not recommend any of the rapid tests for early UTI diagnosis, they showed promise in guiding antimicrobial prescribing and may prove useful with further evidence. The TOUCAN study in England is currently recruiting participants to evaluate point-of-care devices for diagnosing UTIs in primary care. It aims to compare these devices with the current standard testing methods for women who consulting their GP with UTI symptoms [18].
The committee did not recommend culture-based point-of-care tests as they take too long (16-24 hours) to provide results, which would not improve the antibiotic prescribing in primary or community care.
Among the rapid point-of-care tests that show promise in guiding antimicrobial prescribing are Astrego PA-100 analyser with the PA AST panel U-0501 (Sysmex Astrego) and Uriscreen™ (Savyon Diagnostics), both of which have regulatory approval.
The Astrego PA-100 microfluidics-based analyser detects presence of bacteria in the urine specimen in 10-15 minutes. If bacteria are detected, the system assesses the susceptibility of five common urinary pathogens (E. coli, P. mirabilis, K. pneumoniae, S. saprophyticus and E. faecalis) for five antibiotics – amoxicillin / clavulanic acid, ciprofloxacin, fosfomycin, nitrofurantoin, and trimethoprim. Astrego PA-100 is the only rapid test currently assessing the antibiotic susceptibility, and it is being assessed in the TOUCAN study in primary care [19].
Uriscreen is an enzyme-based test that detects the presence of bacterial catalase in a urine sample, detecting both bacteriuria and pyuria, and the results are ready in two minutes [20].
Conclusion
The field of UTI diagnostics is witnessing promising advancements in technologies such as new PCR and FISH arrays, biosensors, and microfluidics. These innovative approaches hold the potential to significantly improve the detection of bacteriuria, enable precise identification of microorganisms, facilitate antimicrobial susceptibility testing (AST), and offer point-of-care testing capabilities. The continued development and research in these areas offer the prospect of more efficient, accurate, and timely UTI diagnosis, ultimately leading to improved patient outcomes.
References
1. EAU. EAU guidelines on urological infections - the guideline. 2023
https://uroweb.org/guidelines/
urological-infections/chapter/the-guideline
2. NHS England. Antimicrobial Stewardship - RightCare UTI. NHS Choices.
https://www.nhsbsa.nhs.uk/access-our-data
-products/epact2/dashboards-and-specifications
/antimicrobial-stewardship-rightcare-uti-focus
-pack-dashboard
3. NICE. New Nice Quality Standard identifies improvements in UTI diagnosis for women. 2023.
https://www.nice.org.uk/news/article/new-nice
-quality-standard-identifies-improvements
-in-uti-diagnosis-for-women
4. Devillé WL, Yzermans JC, van Duijn NP, et al. The urine dipstick test useful to rule out infections. A meta-analysis of the accuracy. BMC Urology 2004;4(1):4.
5. Davenport M, Mach KE, Shortliffe LM, et al. New and developing diagnostic technologies for urinary tract infections. Nature Reviews Urology 2017;14(5):296-310.
6. Kass EH. Pyelonephritis and Bacteriuria. Annals of Internal Medicine 1962;56(1):46.
7. Wilson ML, Gaido L. Laboratory diagnosis of urinary tract infections in adult patients. Clinical Infectious Diseases 2004;38(8):1150-8.
8. Ognenovska S, Mukerjee C, Sanderson-Smith M, et al. Virulence mechanisms of common uropathogens and their intracellular localisation within urothelial cells. Pathogens 2022;11(8):926.
9. Petty LA, Vaughn VM, Flanders SA, et al. Risk factors and outcomes associated with treatment of asymptomatic Bacteriuria in hospitalized patients. JAMA Internal Medicine 2019;179(11):1519.
10. Chronic UTI Info. Broth cultures [Internet]. 2022
https://www.chronicutiinfo.com/
uti_info_accordion/broth-cultures/
11. Xu R, Deebel N, Casals R, et al. A new gold rush: A review of current and developing diagnostic tools for urinary tract infections. Diagnostics 2021;11(3):479.
12. Regenerus labs. Urokey.
https://regeneruslabs.com/
products/microgendx-mens-uti
13. Wu Q, Li Y, Wang M, et al. Fluorescence in situ hybridization rapidly detects three different pathogenic bacteria in urinary tract infection samples. Journal of Microbiological Methods 2010;83(2):175-8.
14. Mach KE, Mohan R, Baron EJ, et al. A biosensor platform for rapid antimicrobial susceptibility testing directly from clinical samples. Journal of Urology 2011;185(1):148-53.
15. Sharma A, Agrawal A, Awasthi KK, et al. Biosensors for diagnosis of urinary tract infections: Advances and future challenges. Materials Letters: X 2021;10:100077.
16. Nanowerk. Mastering the flow in tiny spaces – an overview of microfluidics. 2023
https://www.nanowerk.
com/microfluidics-explained.php
17. NICE. Point-of-care tests for urinary tract infections to improve antimicrobial prescribing: early value assessment. 2023
https://www.nice.org.uk/guidance/
hte7/chapter/1-Recommendations
18. Primary Health Care Clinical Trials unit. TOUCAN: plaTform fOr Uti diagnostiC evAluatioN.
https://www.phctrials.ox.ac.uk/studies/
toucan-platform-for-uti-diagnostic-evaluation
19. Sysmex Astrego. PA-100 AST System.
https://www.sysmex.fr/fileadmin/
media/f100/Business_Lines/POC/SEU
_Astrego_PA-100_flyer_A4_EN_view.pdf
20. Savyon diagnostics. URISCREEN.
https://www.savyondiagnostics.com/
product/uriscreen/
All websites accessed 11 July 2023
Declaration of competing interests: None declared.